#biostripsmedia# #pratheeshpallath#
In this session Bio Strips Media takes through the important
topics of Molecular Basis of Inheritance. It is one of the most important and
scoring topics in Plus Two Biology Exam and NEET Exam.
This lecture covers:
·
Genetic code
·
Translation
GENETIC CODE
Genetic code refers to
the relationship between the sequence of nucleotides in mRNA and the sequence
of amino acids in the polypeptide.
George Gamow, who
suggested that the code must be made up of three bases(triplet codons).
The 64 distinct triplets
determine the 20 amino acids. These triplets are called codons. Any coded
message by these codons is commonly called cryptogram.
Har Gobind Khorana could
synthesise RNA molecules with definite combinations of bases.
Marshal Nierenberg made
a cell free system for protein synthesis that helped in deciphering the code.
Severo Ochoa discovered
enzyme polynucleotide phosphorylase that could polymerise RNA with definite
sequence in template - independent manner.
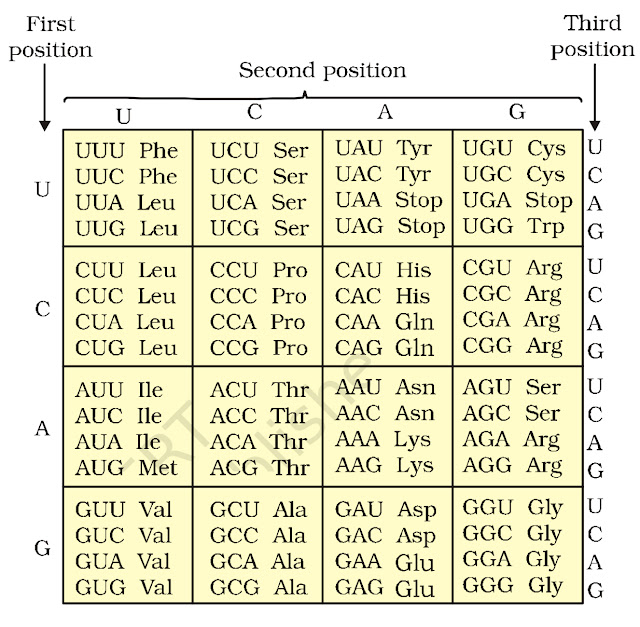
The checkerboard pattern
of genetic code was prepared as given below:
The salient features of
genetic code are given below:
The codons are triplets
and there are 64 codons, 61 codons code for 20 amino acids and 3 codons do not
code for any amino acid, but function as stop/termination codons or nonsense
codons.
Three codons viz UAG
(Amber), UAA (ochre) and UGA (opal) are non sense codons.
Each codons code for
only one amino acid and so the genetic code is unambiguous and specific.
Since some amino acids
are coded by more than one codon, the genetic code is said to be degenerate.
The codons are read in a
contiguous manner in a 5' -’3'’ direction and have no punctuations.
The genetic code is
universal, the codons code for the same amino acid in any organism.
AUG has dual functions
of coding for methionine and acting as initiation codon.
Wobble hypothesis was
given by FHC Crick . According to this, the third nitrogenous base of a
codon is not much significant and Codon is specified by the first two bases.
Hence the same tRNA can recognise more than one codons differing only at third
position.
Translation
It is a process in
living cells in which the genetic information encoded in messenger RNA in the
form of a sequence of nucleotide triplets (codons) is translated into a
sequence of amino acids in a polypeptide chain during protein synthesis.
Ribosomes serve as the
site for protein synthesis hence they are called protein factories. Each
ribosome consists of large and small subunits. the two subunits come
together only at the time of protein synthesis. The phenomena is called
Association . MG 2 + is essential for it. Many ribosomes line up on the mRNA
chain during Protein synthesis. Such a row of active ribosomes is called a
polyribosome or simply a polysome.
Ribosome also acts as a
catalyst (23 s r RNA in bacteria is enzyme ribozyme) for formation of peptide
Bond.
Eukaryotic ribosome has
a groove at the junction of the two subunits. Trom this groove a tunnel extends
through the large subunit and opens into a Canal of the endoplasmic
reticulum. The polypeptides are synthesized in the groove between the two
ribosomal subunits and pass through the Tunnel of the large subunit into the
endoplasmic reticulum. While in the groove the developing polypeptide is protected
from the cellular enzymes. The small subunit forms a cap over the large
subunit. The large subunit attaches to the endoplasmic reticulum by two
glycoproteins named ribophorin 1 and 2.
A ribosome has one
binding site for mRNAi and three binding sites for t RNA. P- site (peptidyl
tRNA site) that holds the tRNA carrying the growing polypeptide chain. A- site
(aminoacyl tRNA site) which holds the tRNA carrying next amino acid to be
added to the chain and E -site (exit site) were discharged tRNAs leave the
ribosome. these sites span across the large subunits of the ribosome. however
the first tRNA amino acid Complex directly enters the P site of the ribosome.
It is a messenger RNA
which brings coded information from DNA and takes part in its translation by
bringing amino acids in a particular sequence during the synthesis of
polypeptide. the codons of mRNA are recognised by anticodons of tRNAs.
Protein synthesis is an
elaborate process and occurs as follows
Activation of amino acids
In this process amino
acid reacts with ATP to form amino acid amp Complex and pyrophosphate. This
complex is called an activated amino acid.
The activated amino acid
attached to the amino acid binding site (3'end) of its specific t RNA. where
its cooh group bonds to 0h group of the terminal base triplet CCA. The process
commonly called as charging of tRNA or aminoacylation of tRNA.
The reaction is
catalysed by the enzyme amino acyl tRNA synthetase.
Initiation of polypeptide chain
The translation of mRNA
begins with the formation of initiation complex. The union of mRNA,a tRNA
bearing the first amino acid of the polypeptide and the two subunits of a
ribosome forms the translation initiation complex.
Proteins called
initiation factors IFs in prokaryotes and eIFs in eukaryotes are necessary to
bring all the components of the translation initiation Complex together. GTP
supplies the energy for the formation of initiation complex.
The small subunit of a
ribosome attaches to mRNA and specific charged initiator tRNA.
In prokaryotes rRNA of
the small subunit base pairs with specific sequence of nucleotides in the mRNAs
leader segment.
in eukaryotes
5Prime cap first tell the small subunit to bind to the mRNAs five Prime end.
The initiator codon
(AUG) which signals the start of translation
The initiator tRNA joins
the initiation codon by its anticodon through hydrogen bonds.
It carries the
amino acid formyl methionine in prokaryotes and methionine in eukaryotes.
Now the large subunit of
ribosome joins the small subunit.
At this stage the
initiator tRNA lies at the P site of the ribosome and the A site is vacant to
let another charged tRNA to enter.
Elongation of polypeptide chain
In the elongation stage
of translation, amino acids are added one by one to the first amino acid. Each
addition requires the help of certain proteins called elongation factors. In
prokaryotes the elongation factors are termed EF-Tu, EF-Ts and EF-G.
An aminoacyl tRNA with
its amino acid enters the ribosome at the A site. It's anticodon reads and
binds to the complementary codon of mRNA by hydrogen bonds.
The amino acid on tRNA
at P site and the newly arrived amino acid at A site join by a peptide Bond. It
is formed between the carboxyl group of first amino acid and the amino group of
the second amino acid. the reaction is catalysed by peptidyl transferase.
Thus the second tRNA
carries a dipeptide and the first t RNA is without amino acid.
Translocation - in
this step the ribosome moves one codon towards the three Prime end of the
mRNA.
This movement shifts the
anti codon of the dipeptidyl tRNA from the A site to the P site and shifts the
deacylated tRNA from the P site to the E site, from where the deacylated tRNA
is released into the cytosol.
The third codon of the
mRNA now lies in the A site and the second codon in the P site.
Now the next cycle
starts. The ribosome moves from codon to codon along the mRNA in the five Prime
to 3 Prime direction.
Amino acids are
added one by one in the sequence of the codon and become joined together to
form polypeptide.
Termination of polypeptide synthesis
Termination is signaled
by the presence of one of three termination codons in the mRNA (UAA, AUG, UGA)
imediately following the final coded amino acid.
When one of the
termination codon comes at A site it does not code for any amino acid and there
is no tRNA molecule for it. as a result the polypeptide synthesis stops.
Three
release factors RF-1, RF-2 and RF-3 contribute in release of the free
polypeptide and last tRNA from the ribosome.
The process is repeated
several times creating polysomes. mRNA has some additional sequences that are
not translated are referred to as untranslated regions (UTRs), required for
efficient translation.


No comments:
Post a Comment